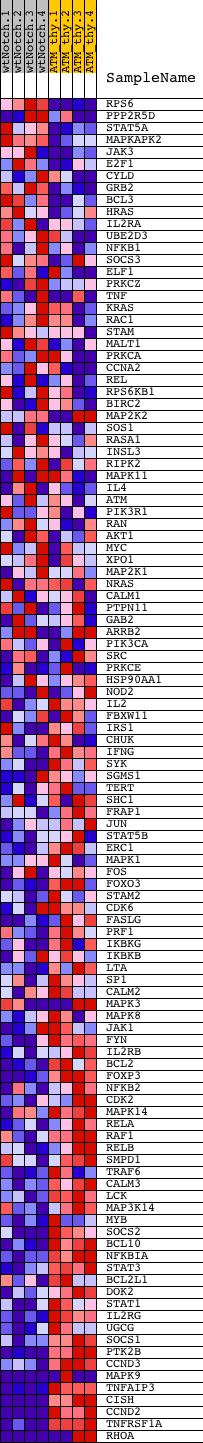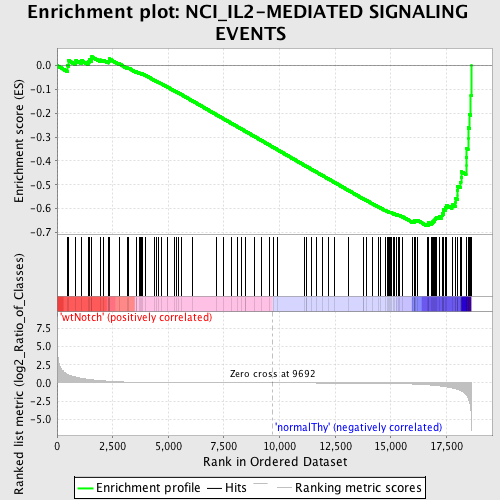
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Set_03_wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy.phenotype_wtNotch_versus_normalThy.cls #wtNotch_versus_normalThy_repos |
| Phenotype | phenotype_wtNotch_versus_normalThy.cls#wtNotch_versus_normalThy_repos |
| Upregulated in class | normalThy |
| GeneSet | NCI_IL2-MEDIATED SIGNALING EVENTS |
| Enrichment Score (ES) | -0.6726594 |
| Normalized Enrichment Score (NES) | -1.649473 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.07817062 |
| FWER p-Value | 0.255 |

| PROBE | DESCRIPTION (from dataset) | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|---|
| 1 | RPS6 | 2060168 | 472 | 1.159 | -0.0003 | No | ||
| 2 | PPP2R5D | 4010156 380408 5550112 5670162 | 524 | 1.084 | 0.0206 | No | ||
| 3 | STAT5A | 2680458 | 844 | 0.819 | 0.0212 | No | ||
| 4 | MAPKAPK2 | 2850500 | 1108 | 0.621 | 0.0205 | No | ||
| 5 | JAK3 | 70347 3290008 | 1394 | 0.500 | 0.0160 | No | ||
| 6 | E2F1 | 5360093 | 1439 | 0.484 | 0.0242 | No | ||
| 7 | CYLD | 6590079 | 1543 | 0.439 | 0.0282 | No | ||
| 8 | GRB2 | 6650398 | 1561 | 0.435 | 0.0367 | No | ||
| 9 | BCL3 | 3990440 | 1936 | 0.326 | 0.0236 | No | ||
| 10 | HRAS | 1980551 | 2087 | 0.286 | 0.0217 | No | ||
| 11 | IL2RA | 6620450 | 2288 | 0.238 | 0.0161 | No | ||
| 12 | UBE2D3 | 3190452 | 2324 | 0.231 | 0.0192 | No | ||
| 13 | NFKB1 | 5420358 | 2339 | 0.229 | 0.0235 | No | ||
| 14 | SOCS3 | 5550563 | 2346 | 0.227 | 0.0281 | No | ||
| 15 | ELF1 | 4780450 | 2784 | 0.154 | 0.0078 | No | ||
| 16 | PRKCZ | 3780279 | 3157 | 0.115 | -0.0098 | No | ||
| 17 | TNF | 6650603 | 3230 | 0.108 | -0.0113 | No | ||
| 18 | KRAS | 2060170 | 3552 | 0.081 | -0.0269 | No | ||
| 19 | RAC1 | 4810687 | 3573 | 0.080 | -0.0262 | No | ||
| 20 | STAM | 3850008 4760441 | 3689 | 0.071 | -0.0309 | No | ||
| 21 | MALT1 | 4670292 | 3761 | 0.066 | -0.0333 | No | ||
| 22 | PRKCA | 6400551 | 3810 | 0.064 | -0.0345 | No | ||
| 23 | CCNA2 | 5290075 | 3843 | 0.062 | -0.0349 | No | ||
| 24 | REL | 360707 | 3994 | 0.055 | -0.0418 | No | ||
| 25 | RPS6KB1 | 1450427 5080110 6200563 | 4372 | 0.041 | -0.0613 | No | ||
| 26 | BIRC2 | 2940435 4730020 | 4458 | 0.039 | -0.0650 | No | ||
| 27 | MAP2K2 | 4590601 | 4558 | 0.036 | -0.0696 | No | ||
| 28 | SOS1 | 7050338 | 4675 | 0.034 | -0.0751 | No | ||
| 29 | RASA1 | 1240315 | 4972 | 0.028 | -0.0905 | No | ||
| 30 | INSL3 | 4150092 | 5287 | 0.023 | -0.1070 | No | ||
| 31 | RIPK2 | 5050072 6290632 | 5362 | 0.023 | -0.1105 | No | ||
| 32 | MAPK11 | 130452 3120440 3610465 | 5433 | 0.022 | -0.1138 | No | ||
| 33 | IL4 | 6020537 | 5571 | 0.020 | -0.1207 | No | ||
| 34 | ATM | 3610110 4050524 | 6095 | 0.015 | -0.1487 | No | ||
| 35 | PIK3R1 | 4730671 | 7181 | 0.009 | -0.2071 | No | ||
| 36 | RAN | 2260446 4590647 | 7473 | 0.008 | -0.2227 | No | ||
| 37 | AKT1 | 5290746 | 7820 | 0.006 | -0.2412 | No | ||
| 38 | MYC | 380541 4670170 | 8113 | 0.005 | -0.2569 | No | ||
| 39 | XPO1 | 540707 | 8292 | 0.005 | -0.2664 | No | ||
| 40 | MAP2K1 | 840739 | 8448 | 0.004 | -0.2747 | No | ||
| 41 | NRAS | 6900577 | 8451 | 0.004 | -0.2747 | No | ||
| 42 | CALM1 | 380128 | 8853 | 0.003 | -0.2963 | No | ||
| 43 | PTPN11 | 2230100 2470180 6100528 | 9170 | 0.002 | -0.3134 | No | ||
| 44 | GAB2 | 1410280 2340520 4280040 | 9538 | 0.000 | -0.3332 | No | ||
| 45 | ARRB2 | 2060441 | 9730 | -0.000 | -0.3435 | No | ||
| 46 | PIK3CA | 6220129 | 9883 | -0.001 | -0.3517 | No | ||
| 47 | SRC | 580132 | 11138 | -0.004 | -0.4194 | No | ||
| 48 | PRKCE | 5700053 | 11223 | -0.005 | -0.4238 | No | ||
| 49 | HSP90AA1 | 4560041 5220133 2120722 | 11225 | -0.005 | -0.4238 | No | ||
| 50 | NOD2 | 2510050 | 11439 | -0.006 | -0.4352 | No | ||
| 51 | IL2 | 1770725 | 11670 | -0.006 | -0.4474 | No | ||
| 52 | FBXW11 | 6450632 | 11943 | -0.008 | -0.4620 | No | ||
| 53 | IRS1 | 1190204 | 12178 | -0.009 | -0.4744 | No | ||
| 54 | CHUK | 7050736 | 12475 | -0.010 | -0.4902 | No | ||
| 55 | IFNG | 5670592 | 13089 | -0.014 | -0.5230 | No | ||
| 56 | SYK | 6940133 | 13759 | -0.022 | -0.5587 | No | ||
| 57 | SGMS1 | 4780195 | 13788 | -0.022 | -0.5597 | No | ||
| 58 | TERT | 6660075 | 13893 | -0.023 | -0.5649 | No | ||
| 59 | SHC1 | 2900731 3170504 6520537 | 14167 | -0.028 | -0.5790 | No | ||
| 60 | FRAP1 | 2850037 6660010 | 14456 | -0.034 | -0.5938 | No | ||
| 61 | JUN | 840170 | 14517 | -0.036 | -0.5963 | No | ||
| 62 | STAT5B | 6200026 | 14758 | -0.044 | -0.6083 | No | ||
| 63 | ERC1 | 1580170 2630717 6110762 6840280 | 14833 | -0.047 | -0.6113 | No | ||
| 64 | MAPK1 | 3190193 6200253 | 14896 | -0.050 | -0.6135 | No | ||
| 65 | FOS | 1850315 | 14961 | -0.053 | -0.6158 | No | ||
| 66 | FOXO3 | 2510484 4480451 | 14977 | -0.054 | -0.6155 | No | ||
| 67 | STAM2 | 730731 2120377 | 15030 | -0.056 | -0.6171 | No | ||
| 68 | CDK6 | 4920253 | 15139 | -0.062 | -0.6215 | No | ||
| 69 | FASLG | 2810044 | 15184 | -0.066 | -0.6225 | No | ||
| 70 | PRF1 | 6660309 | 15276 | -0.071 | -0.6258 | No | ||
| 71 | IKBKG | 3450092 3840377 6590592 | 15354 | -0.077 | -0.6283 | No | ||
| 72 | IKBKB | 6840072 | 15396 | -0.080 | -0.6288 | No | ||
| 73 | LTA | 1740088 | 15537 | -0.093 | -0.6343 | No | ||
| 74 | SP1 | 6590017 | 15961 | -0.142 | -0.6541 | No | ||
| 75 | CALM2 | 6620463 | 16048 | -0.154 | -0.6554 | No | ||
| 76 | MAPK3 | 580161 4780035 | 16052 | -0.154 | -0.6522 | No | ||
| 77 | MAPK8 | 2640195 | 16060 | -0.156 | -0.6492 | No | ||
| 78 | JAK1 | 5910746 | 16124 | -0.165 | -0.6490 | No | ||
| 79 | FYN | 2100468 4760520 4850687 | 16182 | -0.174 | -0.6483 | No | ||
| 80 | IL2RB | 4730072 | 16634 | -0.253 | -0.6671 | Yes | ||
| 81 | BCL2 | 730132 1570736 2470138 3800044 4810037 5690068 5860504 6650164 | 16684 | -0.261 | -0.6641 | Yes | ||
| 82 | FOXP3 | 940707 1850692 | 16687 | -0.262 | -0.6585 | Yes | ||
| 83 | NFKB2 | 2320670 | 16848 | -0.300 | -0.6606 | Yes | ||
| 84 | CDK2 | 130484 2260301 4010088 5050110 | 16881 | -0.308 | -0.6556 | Yes | ||
| 85 | MAPK14 | 5290731 | 16936 | -0.320 | -0.6516 | Yes | ||
| 86 | RELA | 3830075 | 16981 | -0.335 | -0.6467 | Yes | ||
| 87 | RAF1 | 1770600 | 16999 | -0.340 | -0.6402 | Yes | ||
| 88 | RELB | 1400048 | 17053 | -0.356 | -0.6353 | Yes | ||
| 89 | SMPD1 | 1410309 5550717 | 17178 | -0.404 | -0.6332 | Yes | ||
| 90 | TRAF6 | 4810292 6200132 | 17300 | -0.454 | -0.6298 | Yes | ||
| 91 | CALM3 | 3390288 | 17313 | -0.460 | -0.6204 | Yes | ||
| 92 | LCK | 3360142 | 17374 | -0.493 | -0.6129 | Yes | ||
| 93 | MAP3K14 | 5890435 | 17378 | -0.493 | -0.6024 | Yes | ||
| 94 | MYB | 1660494 5860451 6130706 | 17459 | -0.526 | -0.5952 | Yes | ||
| 95 | SOCS2 | 4760692 | 17500 | -0.544 | -0.5855 | Yes | ||
| 96 | BCL10 | 2360397 | 17750 | -0.680 | -0.5842 | Yes | ||
| 97 | NFKBIA | 1570152 | 17914 | -0.797 | -0.5756 | Yes | ||
| 98 | STAT3 | 460040 3710341 | 17921 | -0.802 | -0.5584 | Yes | ||
| 99 | BCL2L1 | 1580452 4200152 5420484 | 17989 | -0.859 | -0.5434 | Yes | ||
| 100 | DOK2 | 5340273 | 17990 | -0.859 | -0.5246 | Yes | ||
| 101 | STAT1 | 6510204 6590553 | 18002 | -0.879 | -0.5061 | Yes | ||
| 102 | IL2RG | 4120273 | 18120 | -1.031 | -0.4900 | Yes | ||
| 103 | UGCG | 3420053 | 18155 | -1.067 | -0.4685 | Yes | ||
| 104 | SOCS1 | 730139 | 18178 | -1.112 | -0.4455 | Yes | ||
| 105 | PTK2B | 4730411 | 18381 | -1.636 | -0.4208 | Yes | ||
| 106 | CCND3 | 380528 730500 | 18383 | -1.645 | -0.3850 | Yes | ||
| 107 | MAPK9 | 2060273 3780209 4070397 | 18415 | -1.759 | -0.3483 | Yes | ||
| 108 | TNFAIP3 | 2900142 | 18472 | -2.065 | -0.3064 | Yes | ||
| 109 | CISH | 840315 | 18476 | -2.102 | -0.2607 | Yes | ||
| 110 | CCND2 | 5340167 | 18548 | -2.713 | -0.2055 | Yes | ||
| 111 | TNFRSF1A | 1090390 6520735 | 18601 | -3.812 | -0.1252 | Yes | ||
| 112 | RHOA | 580142 5900131 5340450 | 18614 | -5.784 | 0.0001 | Yes |